Predicting Life Expectancy using Decision Tree Ensembles
In this post, I will predict life expectancy for the “average” person born in a certain year in one or more countries. I gathered all my data from Gapminder. For the predictor variables, I used birth rates, mortality rates (male, female, under5, and infant), HIV rates, GNI, and Internet usage rates. You can find this data in a compiled csv format (feature_set.csv) here.
There were some missing values of life expectancy in a few of the countries in the earlier years. I imputed these values using linear interpolation. I explored spline interpolation as well, but it did not improve the model’s performance in comparison with linear interpolation.
I considered linear regression, decision trees, random forests, gradient boosting machines, and extra-trees regressors. I also built a validation set for predicting life expectancy for years outside the ranges of the training data provided, specifically for the years 1960 and 2011 to 2015.
The training MSE and validation set MSE are below:
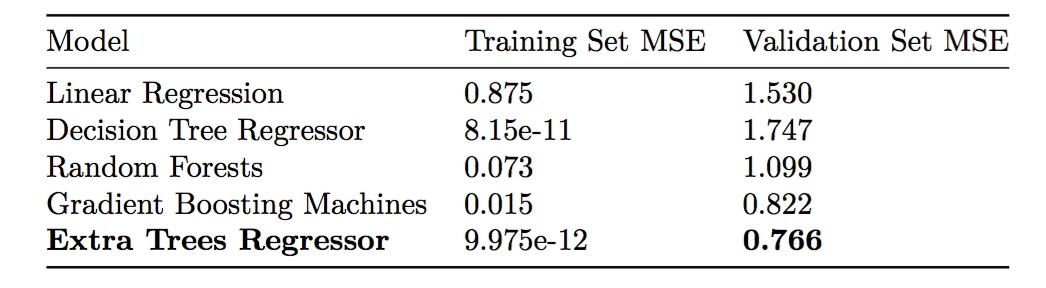
The extra trees regressor performed better than random forests and gradient boosting machines, and the optimal number of trees for this algorithm was 40. As expected, a single decision tree overfits the training data and performs quite poorly on the validation set.
Additional features that I would have liked to include were the total expenditure on health per capita, and pollution estimates. I was unable to acquire accurate estimates for these features, and data was mostly unavailable for the years we considered. However, I believe these features would help predict life expectancy more accurately.
Code
You can find my code here.